SymMap manual
SymMap is an integrative database of traditional Chinese medicine (TCM) with modern medicine (MM). It contains six diverse components, including herb, syndrome, TCM symptom, MM symptom, ingredient, target and disease, which integrating TCM with modern medicine not only by internal molecular mechanism, by also enhanced by symptom mapping.
Users can browse, search and download the six components and their pairwise relationships through the web interface.
- Browse SymMap
- Search in SymMap
- Download data
- Gene functional enrichments
- Detail description of an individual term
- Materials and methods in SymMap
- Citation
1. Browse SymMap
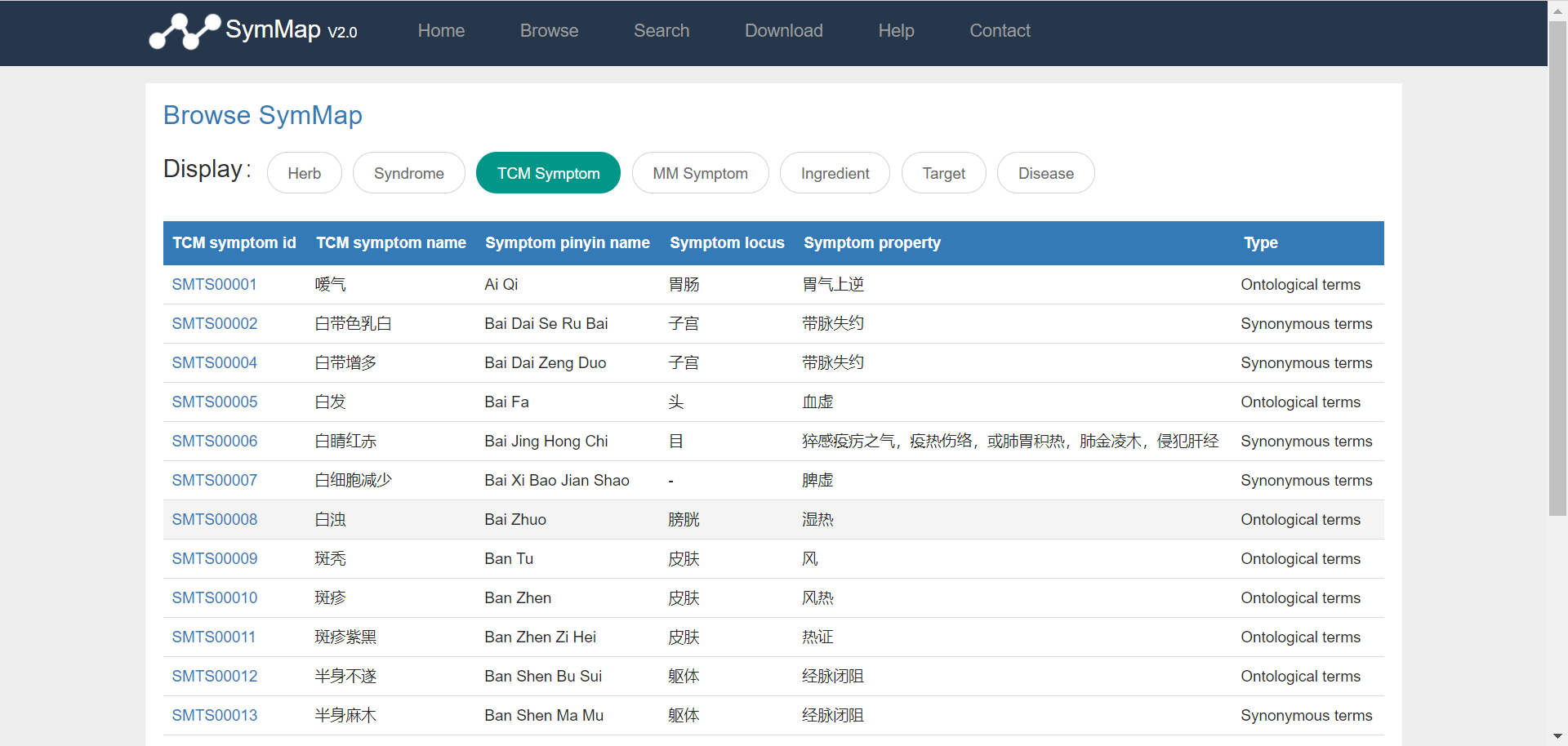
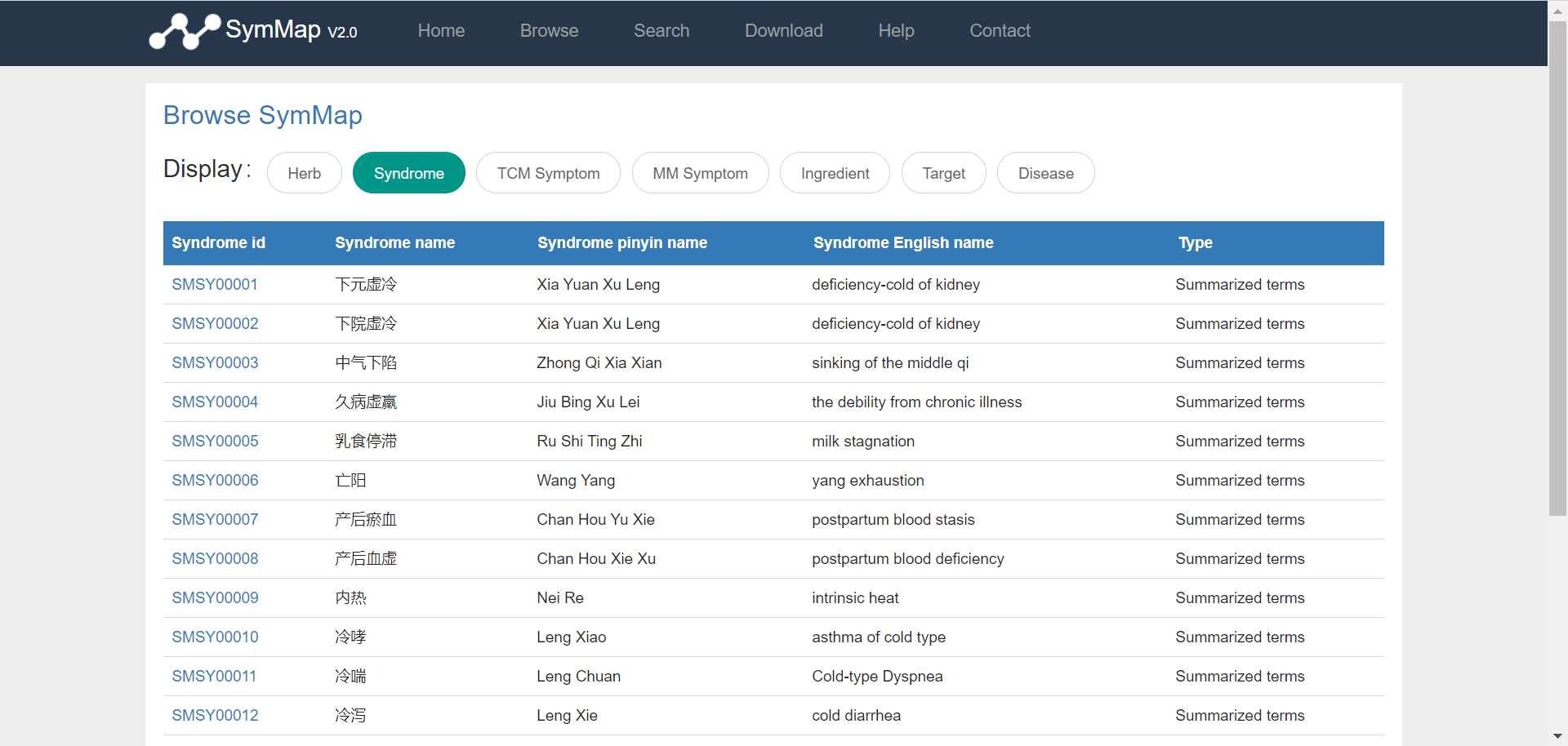
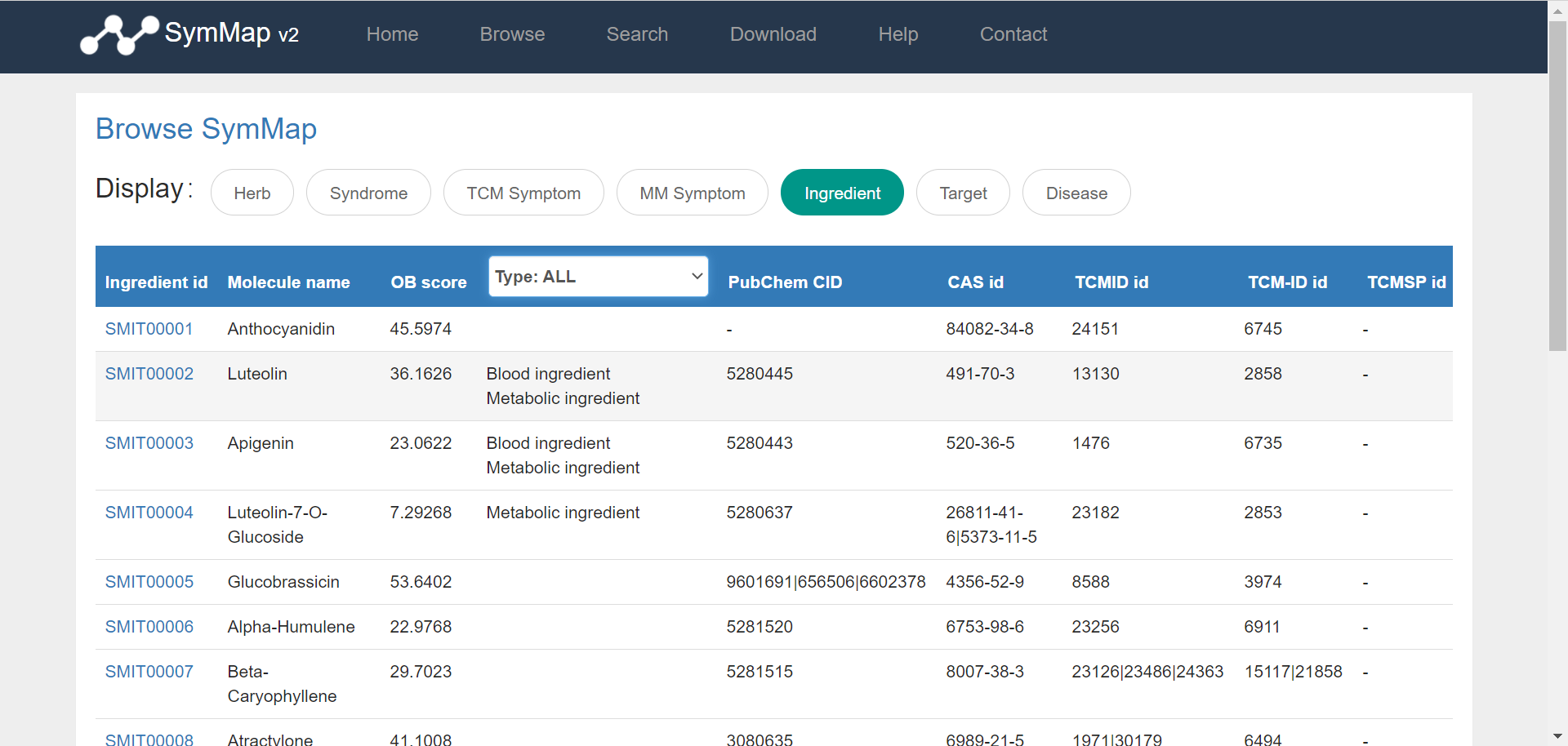
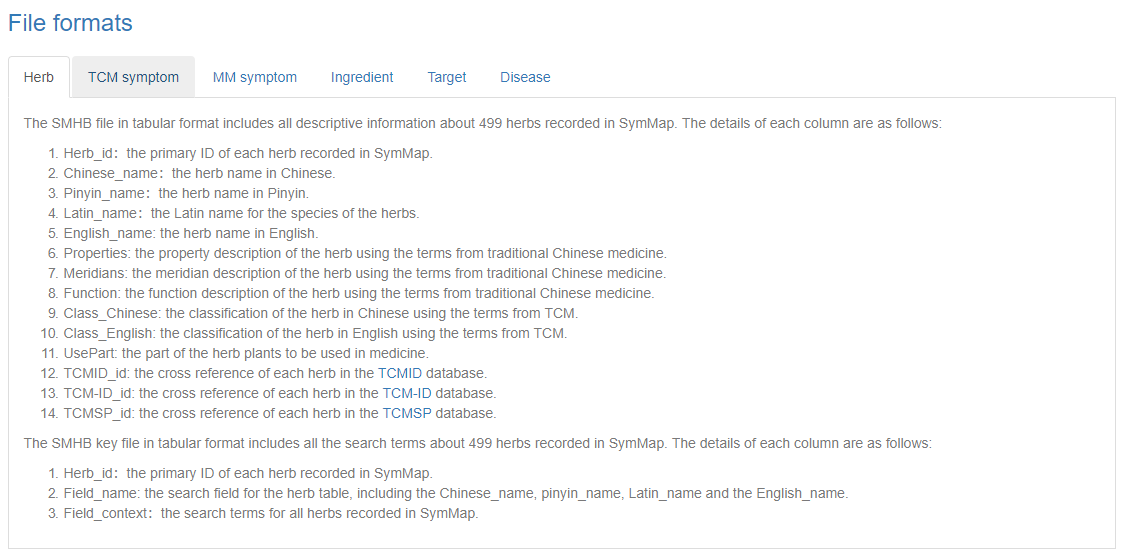
Users can browse the database via clicking the ‘Browse’ button on the homepage, and then access specific data through selecting on the drop down menu in the browse page. Once the users have clicked on a specific component, e.g. TCM symptom (symptom used in traditional Chinese medicine), a summary table about all TCM symptoms is displayed accordingly. Users can get the explanation of the column names in each browse table in the file format section of the download page. Generally, the first column shown in the table is the SymMap ID for each specific term, which contains a hyperlink to its description page in detail. Moreover, users can go through the large datasets by using the pagination menu in the bottom right.
2. Search in SymMap
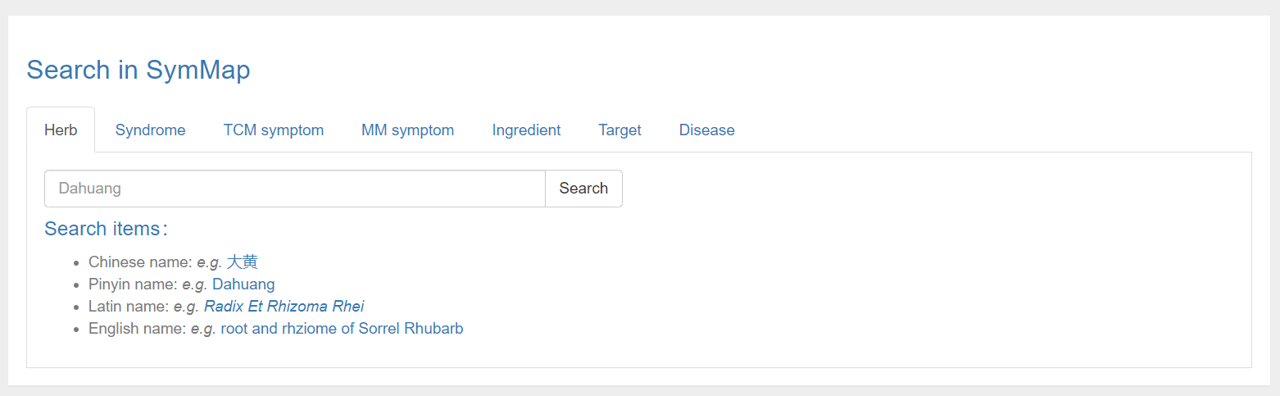
To search in SymMap, users can firstly click the ‘Search’ button on the homepage, and then input and search for an interested query term in the search page. There are six different search boxes provided respectively for each component contained in SymMap. And for each search box, the search items are set distinctively. For example, when users are searching for a specific MM symptom (symptom used in modern medicine), three types of items are permitted to use. The first type is the MM symptom name, e.g. fever. The second type is the external ID of the MM symptom in a widely accepted database, UMLS. The third type includes multiple alias names which are collected from multiple databases for users’ convenience, e.g. pyrexia or temperature raised. All types of allowable search items are displayed behind the search boxes directly. And the explanation of these search items are shown in the file format section of the download page. Users can even download all search items in key files provided by SymMap. Furthermore, the users can select possible matches after only partial query terms input through the autocomplete search functionality conducted in the SymMap.
After searching in SymMap, possible matches for the input query terms are displayed in the lower part of the search page. It is a summary table with the SymMap ID as the first column, which is quite similar to the browse table. Users are encouraged to click the hyperlink on the SymMap ID for detail information.
3. Download data
Information about all the six components in SymMap can be downloaded in the download page. And six key files recording all the search items for each component are also free to download. Moreover, we provided the statistics, the data source, the file sizes for each component in the data table. SymMap automatically conducts gene functional (Pathway and Gene Ontology) enrichment analysis of genes related to these terms for symptoms (TCM and Modern medicine symptoms), diseases, Chinese medicine or ingredients of users' concern. Users can easily download the high-definition figures and complete tables of the enrichment results. More importantly, SymMap can also be used to customize gene collections (i.e., select multiple genes of its own interest) for functional enrichment.

Then we describe the file formats in detail. For each component, the file name is labeled by the prefix of the SymMap ID, which is the abbreviation of the component name. The relationship is as follows:
- SMHB stands for SymMap and herb
- SMYS stands for SymMap and syndrome
- SMTS stands for SymMap and TCM symptom
- SMMS stands for SymMap and MM symptom
- SMIT stands for SymMap and ingredient
- SMTT stands for SymMap and target
- SMDE stands for SymMap and disease
The column names for each file, including the information file and the search key file, are described in detail in the lower part of the download page. All external databases used in SymMap are introduced and linked to its source website respectively.
4. Gene functional enrichments
SymMap automatically conducts gene functional (Pathway and Gene Ontology) enrichment analysis of genes related to these terms for symptoms (TCM and Modern medicine symptoms), diseases, Chinese medicine or ingredients of users' concern. Users can easily download the high-definition figures and complete tables of the enrichment results. More importantly, SymMap can also be used to customize gene collections (i.e., select multiple genes of its own interest) for functional enrichment.
5. Detail description of an individual term
After browse or search in SymMap, users can click the SymMap ID for each specific term to jump onto the detail page, which provided summary information, SymMap network visualization and tables about related components for this specific term orderly.
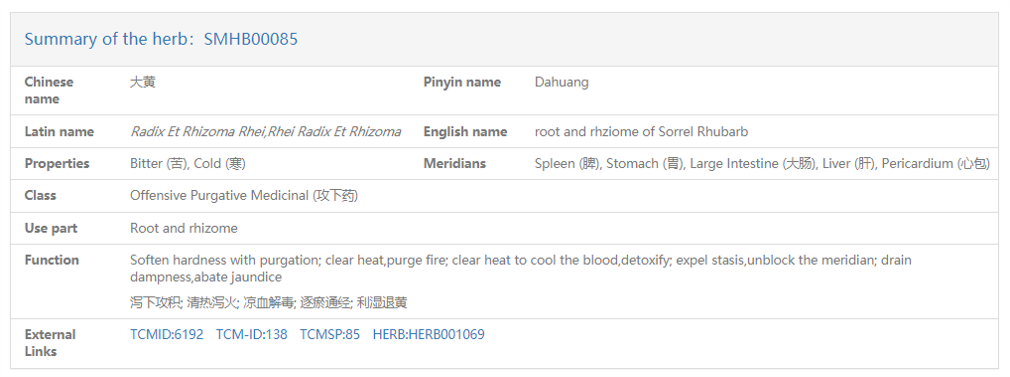
Summary panel displays the descriptive information of the search item. For example, when we stayed on the detail page of the herb, qinghao (青蒿), the summary panel illustrated massive descriptive information on qinghao. All these information comprises five types. The first type is herb_id. The second type is herb names, in Chinese, pinyin, Latin or English. The third type is the descriptions in the TCM terms, for example, property, meridian, and function. The fourth type is the classification of the herb qinghao, in Chinese or in English, as well as the parts used in medicine (use part). And the last type is the IDs of qinghao in other databases about TCM, for example, Herb, TCMID, TCM-ID and TCMSP.
Network panel visualizes all related components for the herb, qinghao. The nodes in the network were colored and placed in different locations according to its component source.
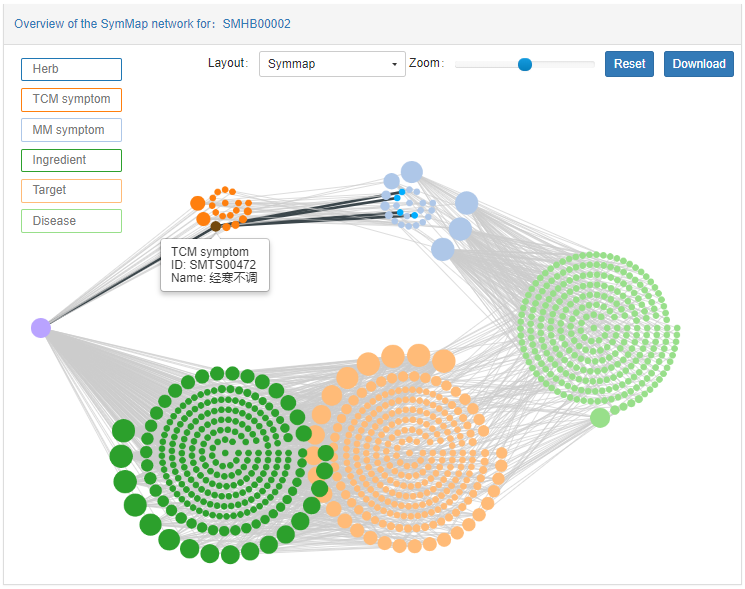
- Herb nodes: dark blue, in the middle-left of the figure
- TCM symptom nodes: dark yellow, in the upper-left of the figure
- MM symptom nodes: light blue, in the upper -right of the figure
- Ingredient nodes: dark green, in the lower-left of the figure
- Target nodes: light yellow, in the lower-right of the figure
- Disease nodes: light green, in the middle-right of the figure
In addition, SymMap also provides two specific networks, namely herb-syndrome-symptom network and ingredient network. The first network contains herbs, syndromes and symptoms, and the network associations among them. The second network consists of quality control components, blood injection components, metabolic components, and their network associations.
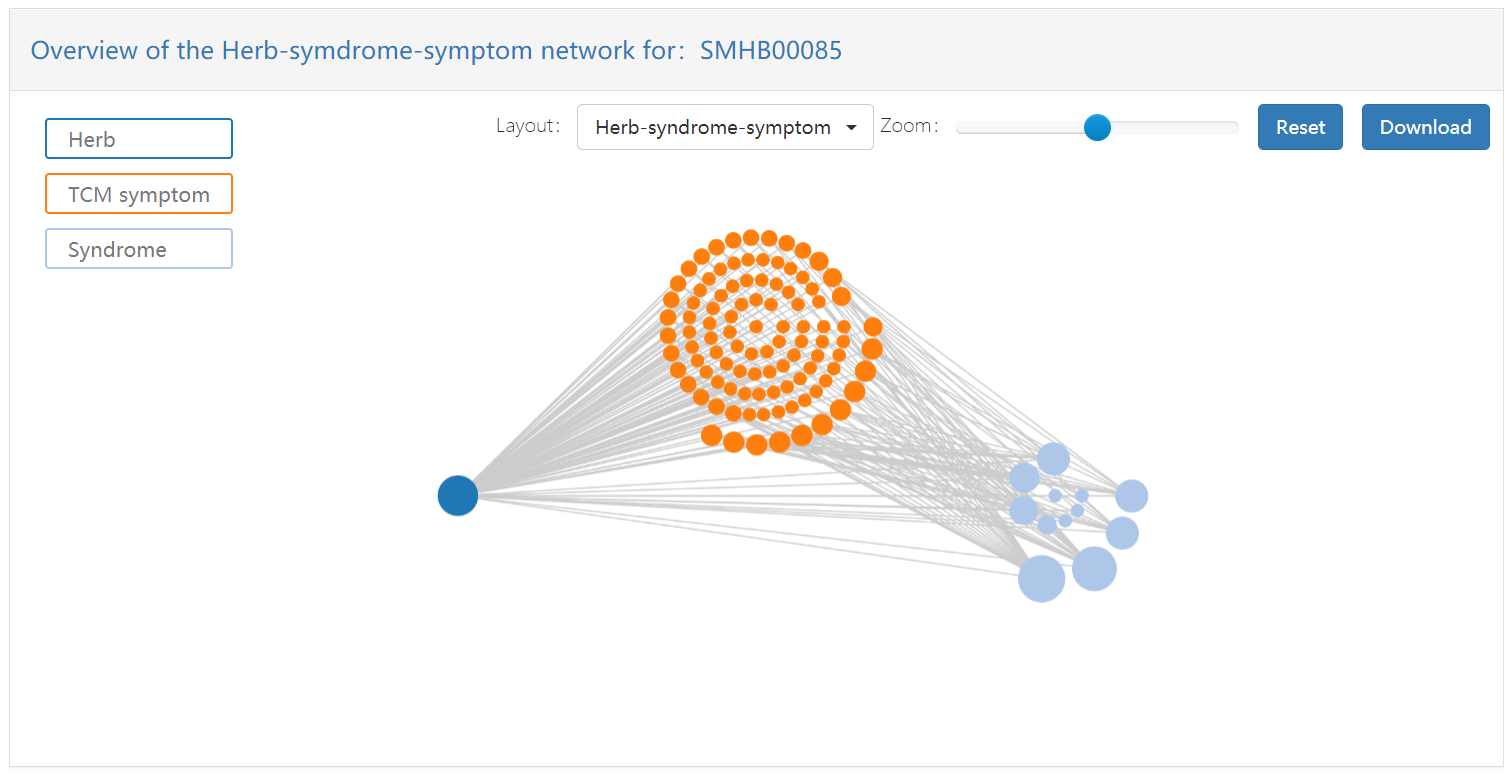
- Herb nodes: dark blue, in the left of the figure
- TCM symptom nodes: dark yellow, in the middle of the figure
- Syndrome nodes: light blue, in the right of the figure
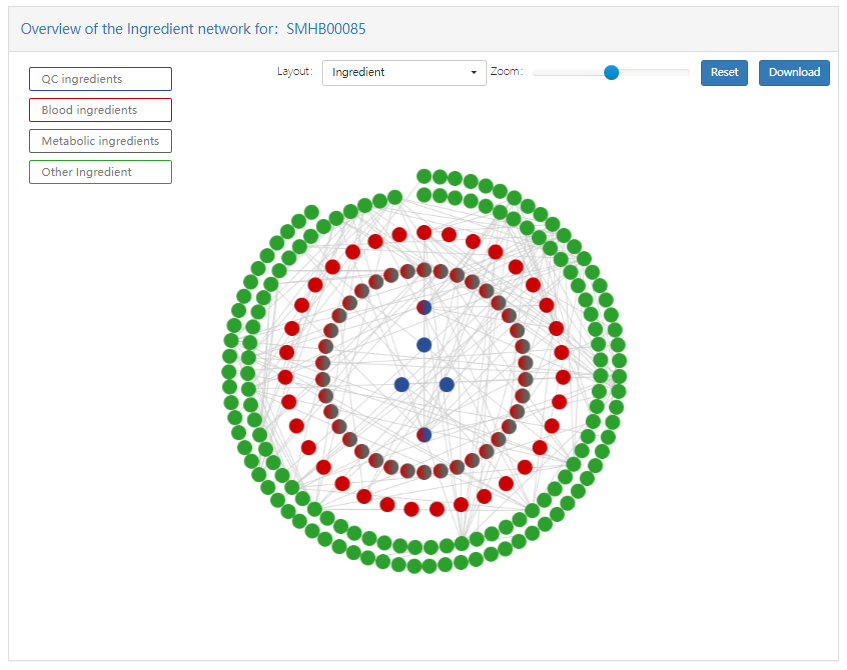
- QC ingredients nodes: blue, in the innermost layer of the figure.
- Metabolic ingredients: dark grey, in the second inner layer of figure.
- Blood ingredients: red, in the second outer layer of figure.
- Other ingredients: green, in the outermost layer of the figure.
For adjacent components, for example, the herb-ingredient relationship, we get the relationship between them according to the manual curation or database mining, or both. For distant components, for example, the TCM symptom-disease relationship, we get their associations by statistical inference on adjacent relationships, i.e. TCM symptom-MM symptom and MM symptom-disease relationships. In brief, the Fisher’s tests and multiple test corrections by using FDR(Bonferroni and BH) method were implemented to remove random indirect associations, in other words, false positives. Only strong indirect associations with a FDR(BH) smaller than 0.05 were retained in the network for accuracy. For more detailed information, please see the method section described in the SymMap paper.
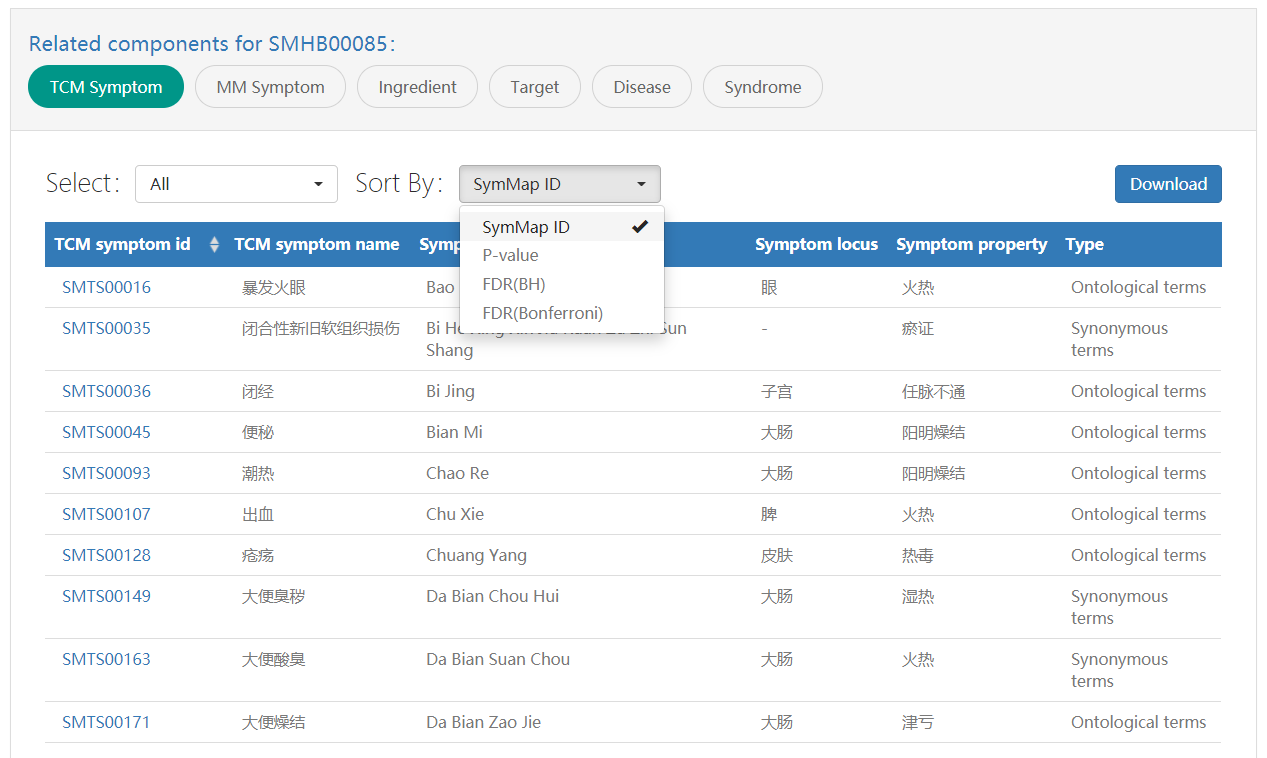
Related component panel is the tabular illustration of the above network. For each specific term, five tables were shown to represent the other five related components, expect for its own component. Users can access a specific component by selecting on the drop down menu. For components having a direct association with the query term, multiple lines in the table shown below are ordered by the SymMap ID ascendingly. For components having an indirect association with the query term, the FDR(Bonferroni and BH) numbers derived from the statistical inference are obtained. For users’ convenience, we provided four set of associations, all, P-value < 0.05 and FDR(Bonferroni and BH) < 0.05. Users can select which level of confidence they would use.
6. Materials and methods in SymMap
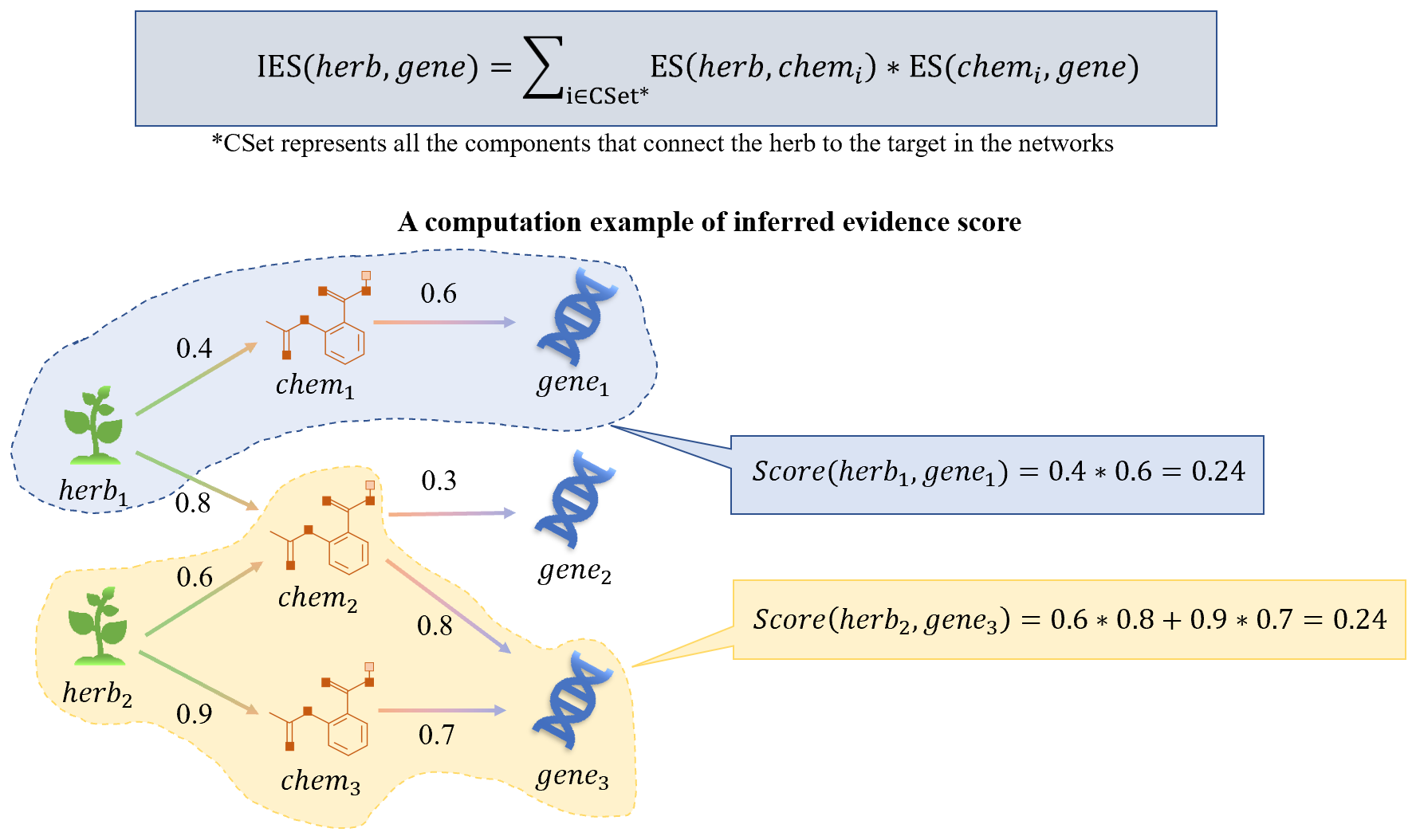
6.1 Inferred evidence score
Inspired by Yang et al. JAMIA 2018 and IEEE JBHI 2019, the IES based on the connection evidence of heterogeneous network has been proposed. First, a heterogeneous network is constructed by integrating all direct relationships among herbs, ingredients, diseases, symptoms and proteins in SymMap. Second, the embedding similarity (ES) between all the direct edges in this network is calculated by the classic network embedding algorithm DeepWalk (Perozzi et al. KDD 2014). Taking the indirect relationship between the herb and the target as an example, all the chemicals (marked by CSet) connecting the herb and the target can be obtained through the ergodic network, and then the IES can be calculated, as follows:
7. Citation
Wu Y#, Zhang F#, Yang K#, Fang S, Bu D, Li H, Sun L, Hu H, Gao K, Wang W, Zhou X*, Zhao Y*, Chen J*. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Research 2018, 47(D1): D1110-D1117.
